Exploratory Data Analysis (EDA) is understanding the data sets by summarizing their main characteristics often plotting them visually. This step is very important especially when we arrive at modeling the data in order to apply Machine learning. In this article, I’ll show you how I did for this!
Introduction
Imaging you are hired as a Senior Data Analyst at Intelligent Insurances Co. The company wants to develop a predictive model that uses vehicle characteristics to accurately predict insurance claim payments. Such a model will allow the company to assess the potential risk that a vehicle represents.
The company puts you in charge of coming up with a solution for this problem and provides you with a historic dataset of previous insurance claims. The claimed amount can be zero or greater than zero and it is given in US dollars.
In this article, I will design my model before conducting some EDA. Let’s get started!
Load Data and Libraries
Load Libraries
import sys
import zipfile
import warnings
import concurrent.futures
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.model_selection import KFold
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import MinMaxScaler
from sklearn.compose import ColumnTransformer
from sklearn.model_selection import StratifiedKFold
from itertools import product
warnings.filterwarnings("ignore")Archive Data
with zipfile.ZipFile("./data/data.zip", 'r') as extractor:
# Print all the contents of the zip file
extractor.printdir()
# Extract all the files
print('Extracting all the files now...')
extractor.extractall(path="./data/")
print('Done!') File Name Modified Size
test.csv 2020-11-12 09:27:10 1989
__MACOSX/._test.csv 2020-11-12 09:27:10 1224
train.csv 2020-10-15 21:32:38 6110914
__MACOSX/._train.csv 2020-10-15 21:32:38 1224
data_dictionary.html 2020-10-15 21:27:32 24739
__MACOSX/._data_dictionary.html 2020-10-15 21:27:32 1280
Extracting all the files now...
Done!Let’s take a quick view.
data = pd.read_csv("./data/train.csv")
data.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 30000 entries, 0 to 29999
Data columns (total 35 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Row_ID 30000 non-null int64
1 Household_ID 30000 non-null int64
2 Vehicle 30000 non-null int64
3 Calendar_Year 30000 non-null int64
4 Model_Year 30000 non-null int64
5 Blind_Make 30000 non-null object
6 Blind_Model 30000 non-null object
7 Blind_Submodel 30000 non-null object
8 Cat1 30000 non-null object
9 Cat2 30000 non-null object
10 Cat3 30000 non-null object
11 Cat4 30000 non-null object
12 Cat5 30000 non-null object
13 Cat6 30000 non-null object
14 Cat7 30000 non-null object
15 Cat8 30000 non-null object
16 Cat9 30000 non-null object
17 Cat10 30000 non-null object
18 Cat11 30000 non-null object
19 Cat12 29948 non-null object
20 OrdCat 30000 non-null object
21 Var1 30000 non-null float64
22 Var2 30000 non-null float64
23 Var3 30000 non-null float64
24 Var4 30000 non-null float64
25 Var5 30000 non-null float64
26 Var6 30000 non-null float64
27 Var7 30000 non-null float64
28 Var8 30000 non-null float64
29 NVCat 30000 non-null object
30 NVVar1 30000 non-null float64
31 NVVar2 30000 non-null float64
32 NVVar3 30000 non-null float64
33 NVVar4 30000 non-null float64
34 Claim_Amount 30000 non-null float64
dtypes: float64(13), int64(5), object(17)
memory usage: 8.0+ MBCheck the types in each columns.
def type_of_col(data, label_col="Claim_Amount", show=True):
df = data.copy()
df = df.drop(label_col, axis=1)
int_features = []
float_features = []
object_features = []
for dtype, feature in zip(df.dtypes, df.columns):
if dtype == 'float64':
float_features.append(feature)
elif dtype == 'int64':
int_features.append(feature)
else:
object_features.append(feature)
if show:
print(f'{len(int_features)} Integer Features : {int_features}\n')
print(f'{len(float_features)} Float Features : {float_features}\n')
print(f'{len(object_features)} Object Features : {object_features}')
return int_features, float_features, object_features
int_features, float_features, object_features = type_of_col(data)Numerical Types
I’ll show you the distribution data if data types are float64 and int64.
df_num = data.select_dtypes(include=['float64', 'int64'])
df_num.hist(figsize=(16, 20), bins=50, xlabelsize=8, ylabelsize=8);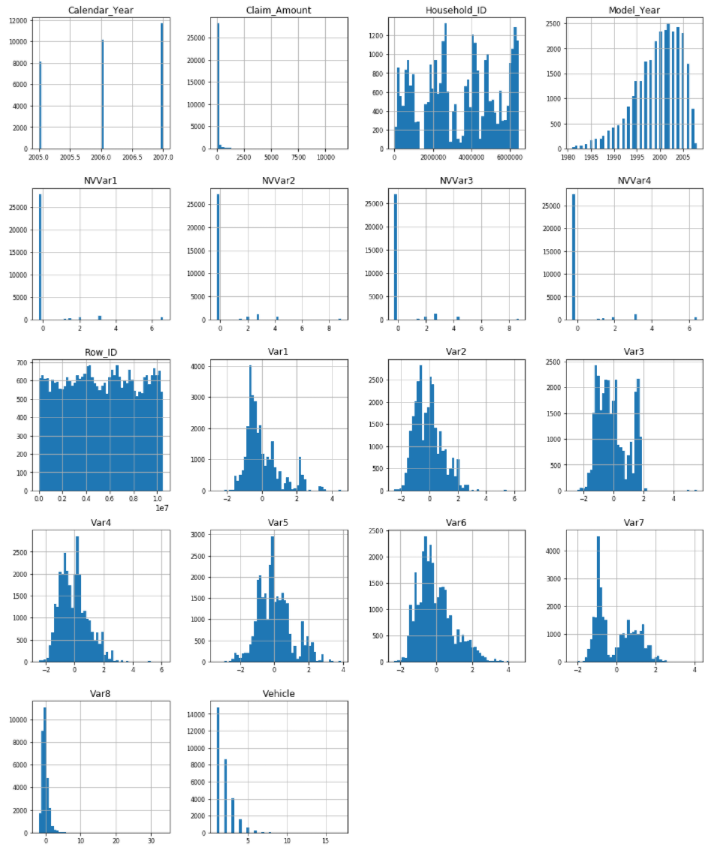
Visualising pairwise relationships in this dataset.
for i in range(0, len(df_num.columns), 5):
sns.pairplot(data=df_num, x_vars=df_num.columns[i:i+5], y_vars=['Claim_Amount'])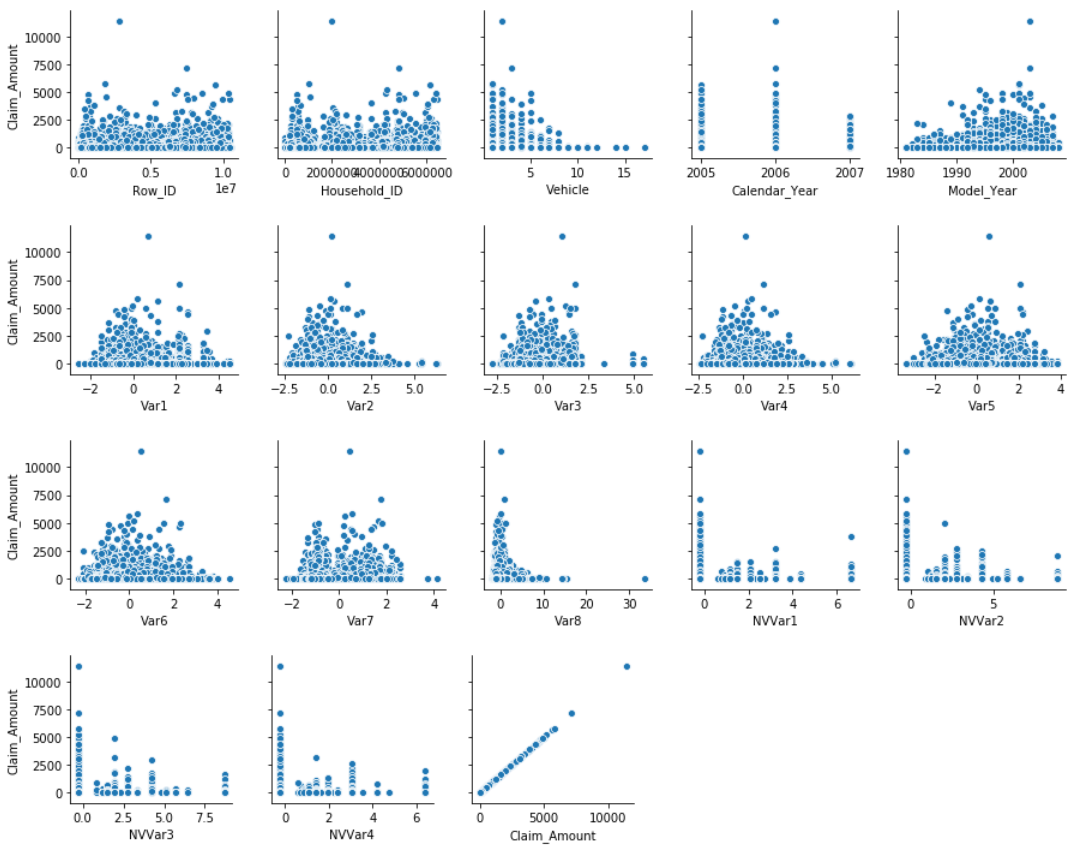
Review heatmaps among numerical types of data.
corr = df_num.drop('Claim_Amount', axis=1).corr()
plt.figure(figsize=(12, 10))
sns.heatmap(
corr[(corr >= 0.5) | (corr <= -0.4)],
cmap='viridis', vmax=1.0, vmin=-1.0, linewidths=0.1,
annot=True, annot_kws={"size": 8}, square=True);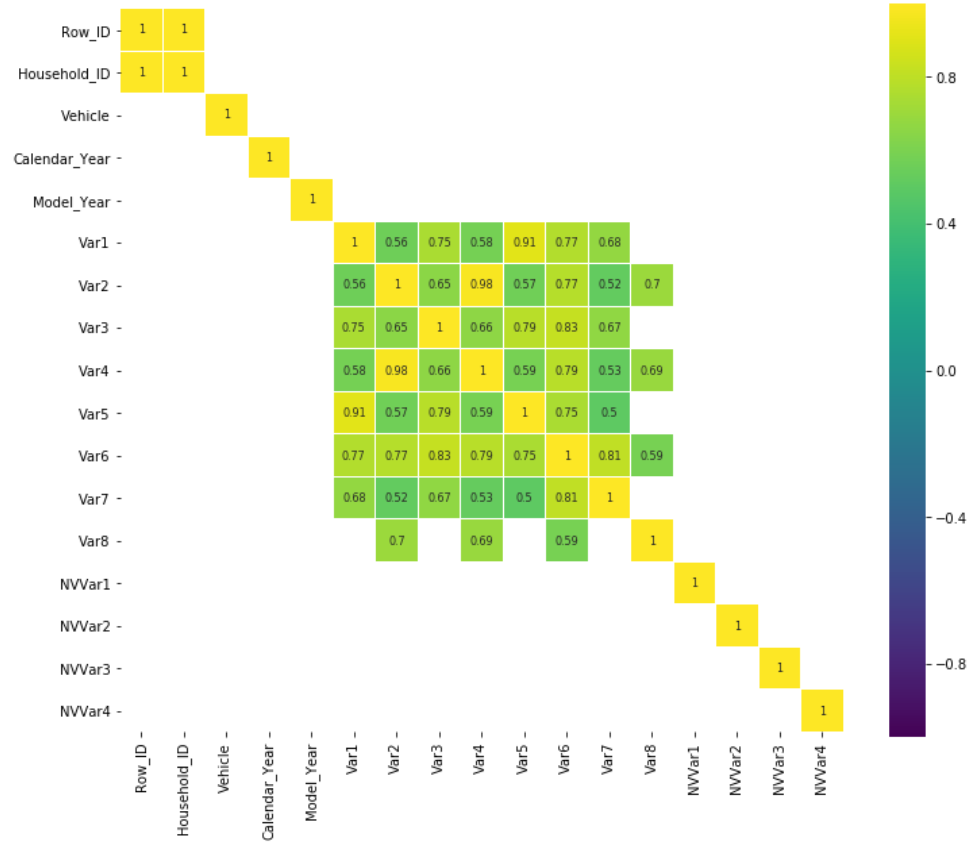
Categorical Types
While visualising categorical types of data, I saw there are some “?” in some features.
df_not_num = data.select_dtypes(include=['O'])
fig, axes = plt.subplots(round(len(df_not_num.columns) / 3), 3, figsize=(12, 20))
for i, ax in enumerate(fig.axes):
if i < len(df_not_num.columns):
ax.set_xticklabels(ax.xaxis.get_majorticklabels(), rotation=45)
sns.countplot(x=df_not_num.columns[i], alpha=0.7, data=df_not_num, ax=ax)
fig.tight_layout()
plt.show()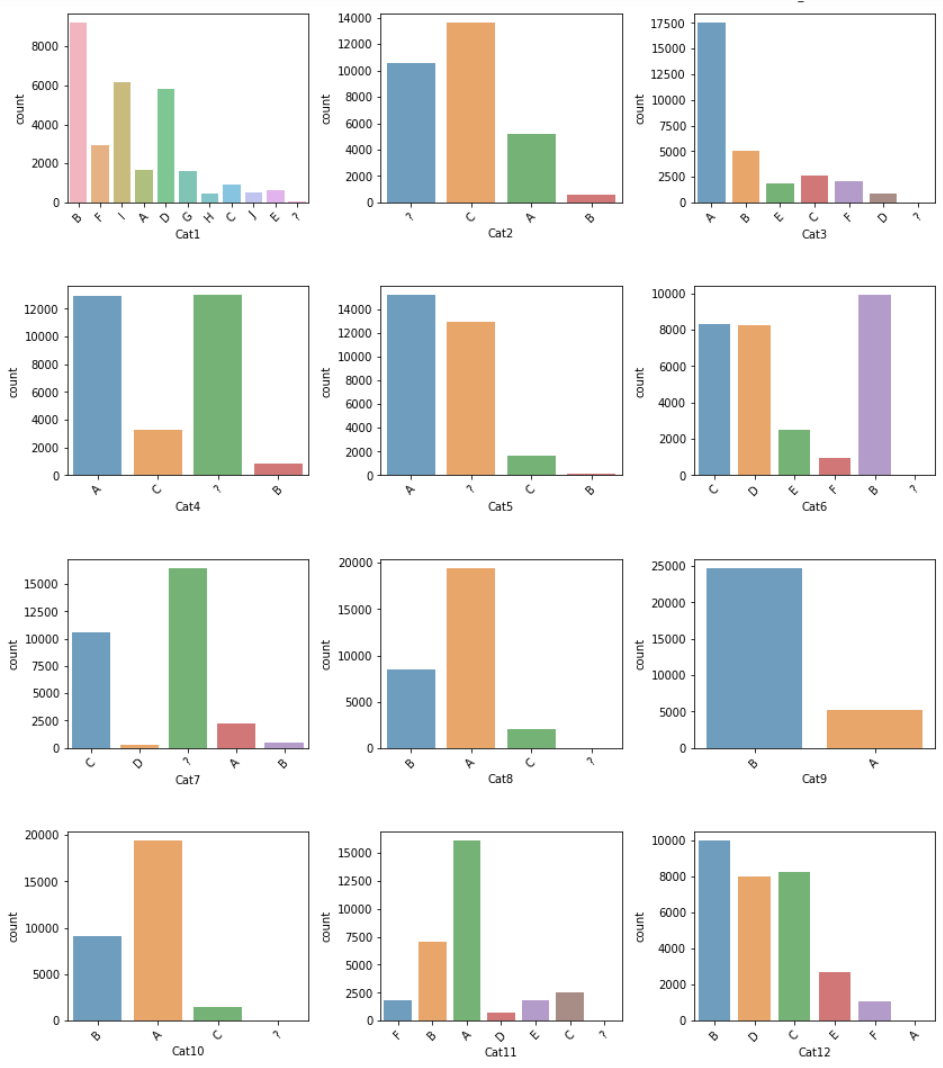
Feature Selection
Feature Selection is the process where you automatically or manually select those features which contribute most to your prediction variable or output in which you are interested in.
| Feature \ Label | Continuous | Categorical |
|---|---|---|
| Continuous | Pearson’s Correlation | LDA |
| Categorical | ANOVA | Chi-Square |
Featrue Selection using Chi-Square Test
The Chi-Square test of independence is a statistical test to determine if there is a significant relationship between 2 categorical variables. In simple words, the Chi-Square statistic will test whether there is a significant difference in the observed vs the expected frequencies of both variables.
import scipy.stats as stats
from scipy.stats import chi2_contingency
class ChiSquare:
""""
H0: No association between two variables.
H1: There is evidence to suggest there is an association between two variables.
"""
def __init__(self, data):
self.data = data
self.p_value = None
self.chi2 = None
self.dof = None
self.data_observed = None
self.data_expected = None
self.important_features = []
self.unimportant_features = []
def print_result(self, col, alpha=0.05):
if self.p_value < alpha:
# Reject null hypothesis H0
print(f"{col} is an IMPORTANT feature.")
else:
# Accept null hypothesis H0
print(f"{col} is NOT an IMPORTANT feature.")
def get_result(self, col, alpha=0.05):
if self.p_value < alpha:
# Reject null hypothesis H0
self.important_features.append(col)
else:
# Accept null hypothesis H0
self.unimportant_features.append(col)
def get_important_features(self):
return self.important_features
def get_unimportant_features(self):
return self.unimportant_features
def test(self, col_features, col_y, alpha=0.05):
for col_x in col_features:
X = self.data[col_x].astype(str)
y = self.data[col_y].apply(lambda label: 0 if label==0.0 else 1).astype(str)
self.data_observed = pd.crosstab(y, X)
chi2, p_value, dof, expected = chi2_contingency(self.data_observed.values)
self.chi2 = chi2
self.p_value = p_value
self.dof = dof
self.data_expected = pd.DataFrame(
expected,
columns=self.data_observed.columns,
index=self.data_observed.index)
self.get_result(col_x, alpha)
self.print_result(col_x, alpha)Chi-Square Test for Categorical Features.
chi_test = ChiSquare(data)
test_cols = df_not_num.columns.tolist()
chi_test.test(test_cols, "Claim_Amount")
important_cat_features = chi_test.get_important_features()
unimportant_cat_features = chi_test.get_unimportant_features()Blind_Make is an IMPORTANT feature.
Blind_Model is an IMPORTANT feature.
Blind_Submodel is NOT an IMPORTANT feature.
Cat1 is an IMPORTANT feature.
Cat2 is an IMPORTANT feature.
Cat3 is an IMPORTANT feature.
Cat4 is an IMPORTANT feature.
Cat5 is an IMPORTANT feature.
Cat6 is an IMPORTANT feature.
Cat7 is an IMPORTANT feature.
Cat8 is NOT an IMPORTANT feature.
Cat9 is an IMPORTANT feature.
Cat10 is NOT an IMPORTANT feature.
Cat11 is NOT an IMPORTANT feature.
Cat12 is NOT an IMPORTANT feature.
OrdCat is an IMPORTANT feature.
NVCat is an IMPORTANT feature.You can find out that ‘Blind_Make’, ‘Blind_Model’, ‘Cat1’, ‘Cat2’, ‘Cat3’, ‘Cat4’, ‘Cat5’, ‘Cat6’, ‘Cat7’, ‘Cat9’, ‘OrdCat’, ‘NVCat’ are important features, and ‘Blind_Submodel’, ‘Cat8’, ‘Cat10’, ‘Cat11’, ‘Cat12’ are unimportant features by Chi-Square Test.
Feature Selection using XGBoost
import xgboost as xgb
from xgboost import XGBClassifier
from xgboost import plot_importance
from sklearn.metrics import f1_score
def model(X_train, y_train, n_splits=3):
scores=[]
params = {
'colsample_bytree': 0.8,
'learning_rate': 0.08,
'max_depth': 10,
'subsample': 1,
'objective': 'multi:softprob',
'num_class': 2,
'eval_metric': 'mlogloss',
'min_child_weight': 3,
'gamma': 0.25,
}
kf = StratifiedKFold(n_splits=n_splits, shuffle=True, random_state=42)
for train_index, val_index in kf.split(X_train, y_train):
train_X = X_train.iloc[train_index]
val_X = X_train.iloc[val_index]
train_y = y_train[train_index]
val_y = y_train[val_index]
xgb_train = xgb.DMatrix(train_X, train_y)
xgb_eval = xgb.DMatrix(val_X, val_y)
xgb_model = xgb.train(
params,
xgb_train,
num_boost_round=1000,
evals=[(xgb_train, 'train'), (xgb_eval, 'val')],
verbose_eval=False,
early_stopping_rounds=20
)
val_X = xgb.DMatrix(val_X)
pred_val = [np.argmax(x) for x in xgb_model.predict(val_X)]
score = f1_score(pred_val, val_y)
scores.append(score)
print('F1 score: ', score)
return xgb_model
num_feature = [
'Vehicle', 'Calendar_Year', 'Model_Year', 'Var1', 'Var2', 'Var3',
'Var4', 'Var5', 'Var6', 'Var7', 'Var8', 'NVVar1', 'NVVar2', 'NVVar3', 'NVVar4']
xgb_model = model(data[num_feature], (data.Claim_Amount!=0).astype(int), n_splits=5)
fig, ax = plt.subplots(figsize=(10, 10))
xgb.plot_importance(xgb_model, max_num_features=50, height=0.5, ax=ax, importance_type='gain',show_values=False)
plt.show()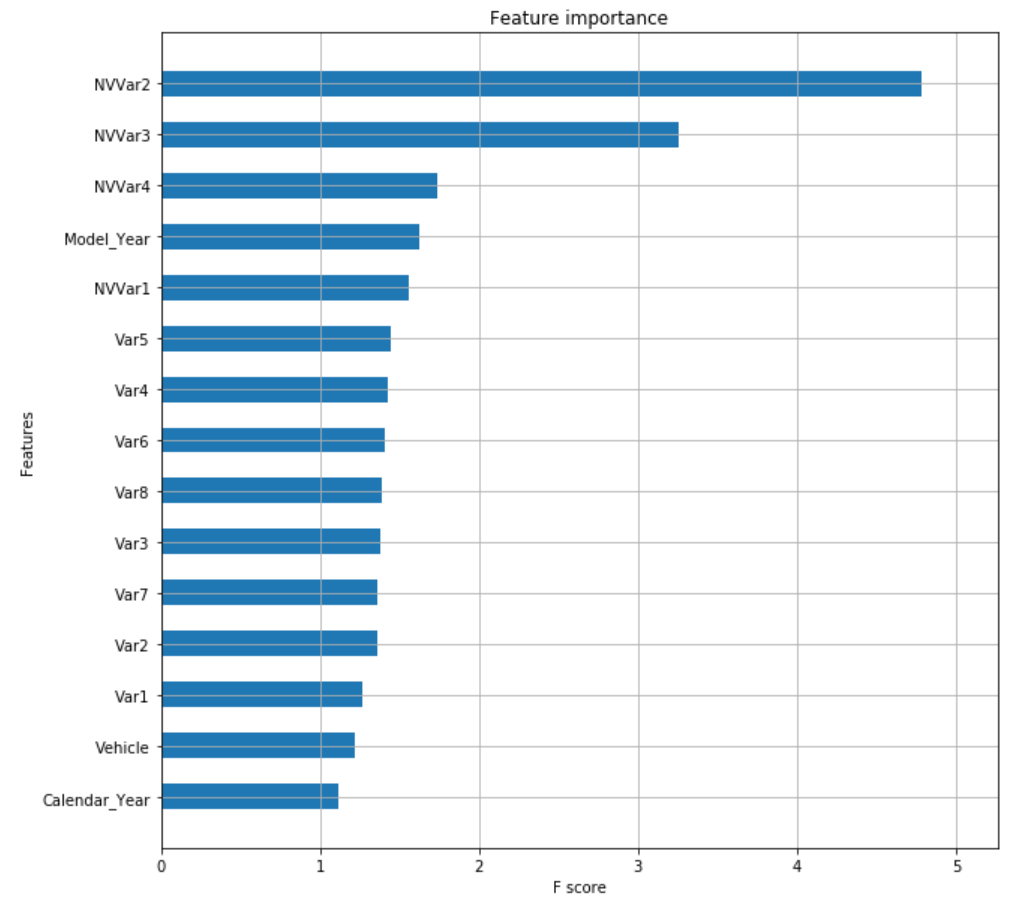
Missing Data
In this dataset, missing data is represented by a ‘?’ or a missing value. Therefore, it should start with tackling missing data. First, fill all None and missing data with np.nan. Second, replace ‘?’ by np.nan. Finally, plot a heatmap to get a view over the dataset.
def plot_missing_value_heatmap(data):
plt.figure(figsize=(15, 10))
sns.heatmap(data.isnull(), cbar=True, cmap=sns.color_palette("cubehelix"))
plt.title("Missing Values Heatmap", fontdict={'family': 'serif', 'weight': 'normal', 'size': 16,})
plt.show()
data_pre = data.copy()
data_pre.fillna(value=np.nan, inplace=True)
for col in object_features:
data_pre.loc[:, col] = data_pre.loc[:, col].replace(to_replace='?',value=np.nan)
data_pre.loc[:, col] = data_pre.loc[:, col].replace(to_replace='""',value=np.nan)
plot_missing_value_heatmap(data_pre)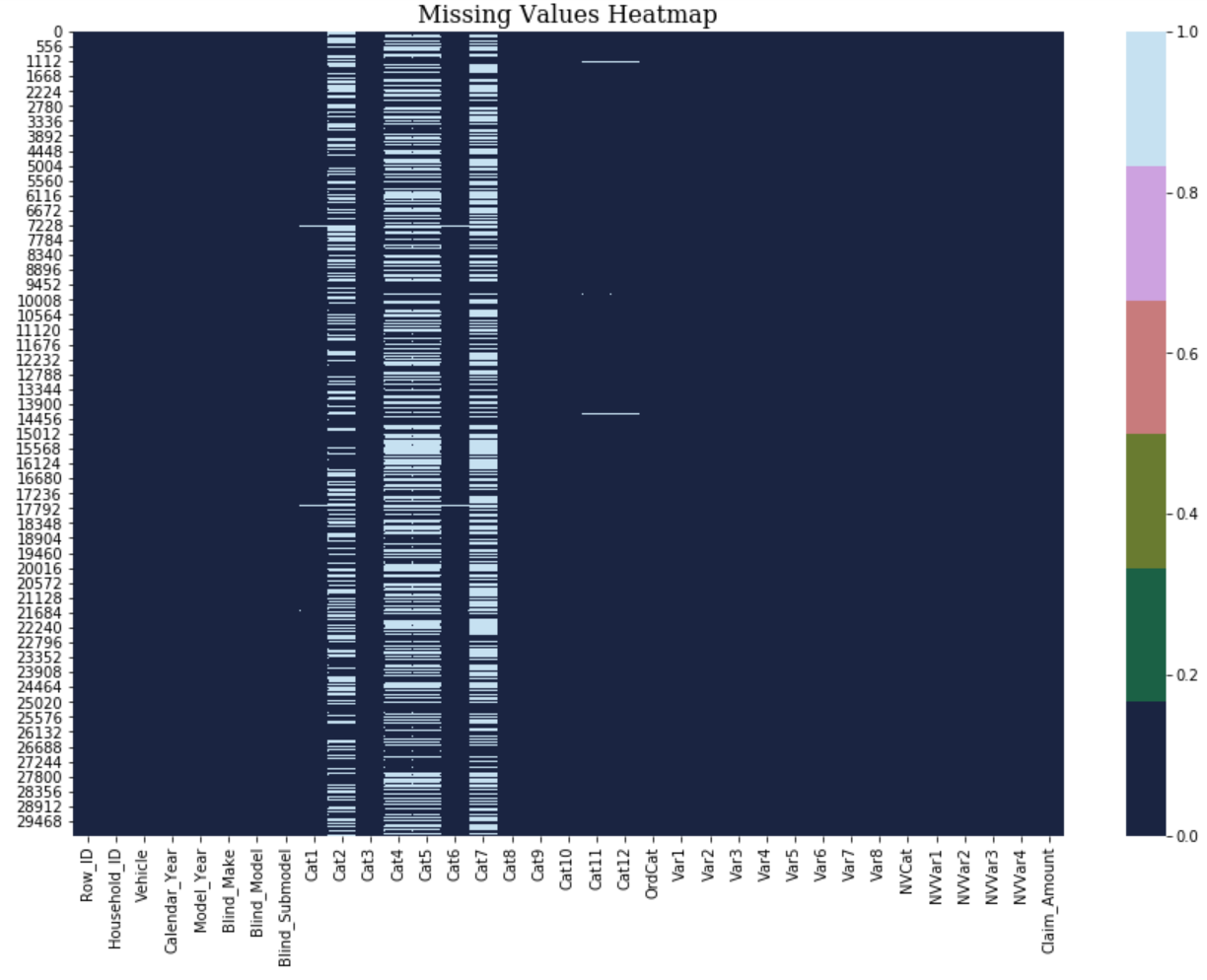
There are too many missing values in columns below:
- Cat2
- Cat4
- Cat5
- Cat7
So I just drop those columns.
# Split data into features and label
data_pre_feature1 = data_pre.drop('Claim_Amount', axis=1)
data_pre_label = data_pre['Claim_Amount']
# Convert int dtype into float dtype
for col in int_features:
data_pre_feature1[col] = data_pre_feature1[col].astype('float64')
data_pre_feature1 = data_pre_feature1.drop(["Cat2", "Cat4", "Cat5", "Cat7"], axis=1)
plot_missing_value_heatmap(data_pre_feature1)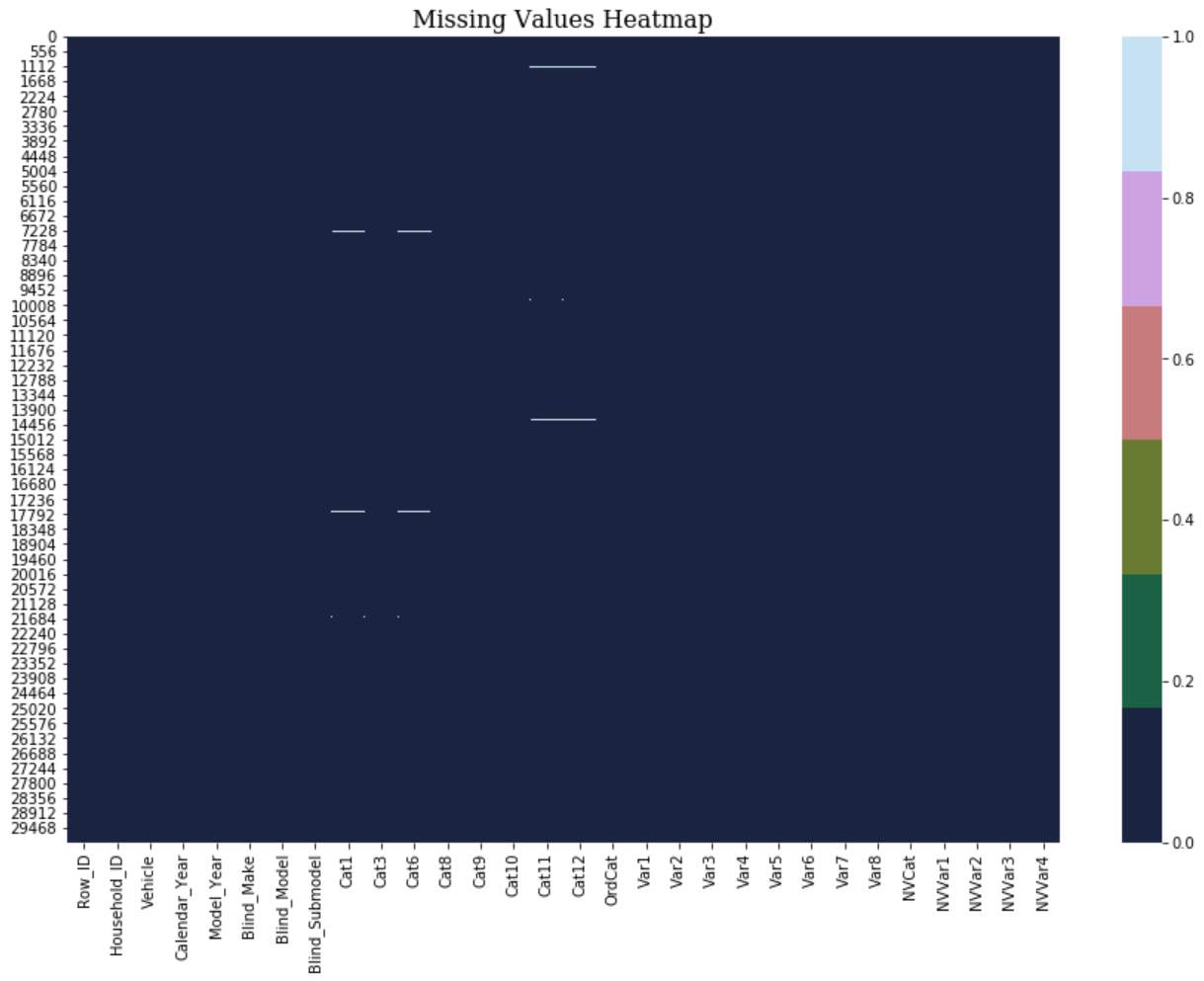
It’s quite good after removing these four columns! Therefore, for those features (“Cat2”, “Cat4”, “Cat5”, “Cat7”) which have more than 90% of missing value, I chose dropping them instead of filling . Rest of the featuers I chose using forward filling method from pandas library.
Features Transformation
I utilised different transformation methods for different data types.
- ‘Blind_Make’, ‘Blind_Model’, ‘Blind_Submodel’:
MeanEncoder()for high-cardinality categorical data. - ‘Row_ID’, ‘Household_ID’, ‘Vehicle’, ‘Calendar_Year’, ‘Model_Year’, ‘Var1’, ‘Var2’, ‘Var3’, ‘Var4’, ‘Var5’, ‘Var6’, ‘Var7’, ‘Var8’, ‘NVVar1’, ‘NVVar2’, ‘NVVar3’, ‘NVVar4’:
MinMaxScaler()for numerical dat. - ‘Cat1’, ‘Cat3’, ‘Cat6’, ‘Cat8’, ‘Cat9’, ‘Cat10’, ‘Cat11’, ‘Cat12’, ‘OrdCat’, ‘NVCat’:
OneHotEncoder()for categorical data (but in smaller dimension).
data_pre_feature2 = data_pre_feature1.fillna(method='ffill')
num_feature = [
'Row_ID', 'Household_ID', 'Vehicle', 'Calendar_Year', 'Model_Year', 'Var1', 'Var2', 'Var3',
'Var4', 'Var5', 'Var6', 'Var7', 'Var8', 'NVVar1', 'NVVar2', 'NVVar3', 'NVVar4']
cat_feature = [
'Cat1', 'Cat3', 'Cat6', 'Cat8', 'Cat9', 'Cat10', 'Cat11', 'Cat12', 'OrdCat', 'NVCat']
full_transform = ColumnTransformer([
("num", StandardScaler(), num_feature),
("cat", OneHotEncoder(), cat_feature)
])
data_pre_feature3 = full_transform.fit_transform(data_pre_feature2)Mean Encoder
Mean Encoding is a simple preprocessing scheme for high-cardinality categorical data that allows this class of attributes to be used in predictive models such as neural networks, linear and logistic regression. The proposed method is based on a well-established statistical method (empirical Bayes) that is straightforward to implement as an in-database procedure. Furthermore, for categorical attributes with an inherent hierarchical structure, like ZIP codes, the preprocessing scheme can directly leverage the hierarchy by blending statistics at the various levels of aggregation.
I made this MeanEncoder() into sklearn-compatible class object.
class MeanEncoder(TransformerMixin, BaseEstimator):
"""
http://helios.mm.di.uoa.gr/~rouvas/ssi/sigkdd/sigkdd.vol3.1/barreca.pdf
"""
def __init__(self, cat_features, cv=10, target_type='classification', weight_func=None, k=2, f=1):
self.cat_features = cat_features
self.cv = cv
self.k = k
self.f = f
self.learned_stats = {}
if target_type == 'classification':
self.target_type = target_type
self.target_values = []
elif target_type == 'regression':
self.target_type = 'regression'
self.target_values = None
else:
print("Label type could only be 'classification' or 'regression'.")
# Calculate smoothing factor: 1 / (1 + np.exp(- (counts - min_samples_leaf) / smoothing_slope))
if isinstance(weight_func, dict):
self.weight_func = eval(
'lambda x: 1 / (1 + np.exp(-(x-k)/f))', dict(weight_func, np=np, k=k, f=f))
elif callable(weight_func):
self.weight_func = weight_func
else:
self.weight_func = lambda x: 1 / (1 + np.exp(-(x-k)/f))
# For training dataset
def fit_transform(self, X, y):
X_new = X.copy()
if self.target_type == 'classification':
skf = StratifiedKFold(self.cv)
else:
skf = KFold(self.cv)
# Categorical label
if self.target_type == 'classification':
self.target_values = sorted(set(y))
self.learned_stats = {'{}_pred_{}'.format(variable, target): [] for variable, target in
product(self.cat_features, self.target_values)}
for variable, target in product(self.cat_features, self.target_values):
nf_name = '{}_pred_{}'.format(variable, target)
X_new.loc[:, nf_name] = np.nan
for large_ind, small_ind in skf.split(y, y):
nf_large, nf_small, prior, col_avg_y = MeanEncoder.mean_encode_blended(
X_new.iloc[large_ind],
y.iloc[large_ind],
X_new.iloc[small_ind],
variable,
target,
self.weight_func)
X_new.iloc[small_ind, -1] = nf_small
self.learned_stats[nf_name].append((prior, col_avg_y))
# Continuous label
else:
self.learned_stats = {'{}_pred'.format(variable): [] for variable in self.cat_features}
for variable in self.cat_features:
nf_name = '{}_pred'.format(variable)
X_new.loc[:, nf_name] = np.nan
for large_ind, small_ind in skf.split(y, y):
nf_large, nf_small, prior, col_avg_y = MeanEncoder.mean_encode_blended(
X_new.iloc[large_ind],
y.iloc[large_ind],
X_new.iloc[small_ind],
variable,
None,
self.weight_func)
X_new.iloc[small_ind, -1] = nf_small
self.learned_stats[nf_name].append((prior, col_avg_y))
X_new = X_new.drop(self.cat_features, axis=1)
X_new.columns = self.cat_features
return X_new
# For testing dataset
def transform(self, X):
X_new = X.copy()
# Categorical label
if self.target_type == 'classification':
for variable, target in product(self.cat_features, self.target_values):
nf_name = '{}_pred_{}'.format(variable, target)
X_new[nf_name] = 0
for prior, col_avg_y in self.learned_stats[nf_name]:
X_new[nf_name] += X_new[[variable]].join(
col_avg_y, on=variable).fillna(prior, inplace=False)[
nf_name]
X_new[nf_name] /= self.cv
# Continuous label
else:
for variable in self.cat_features:
nf_name = '{}_pred'.format(variable)
X_new[nf_name] = 0
for prior, col_avg_y in self.learned_stats[nf_name]:
X_new[nf_name] += X_new[[variable]].join(
col_avg_y, on=variable).fillna(prior, inplace=False)[
nf_name]
X_new[nf_name] /= self.cv
X_new = X_new.drop(self.cat_features, axis=1)
X_new.columns = self.cat_features
return X_new
# Prior probability and posterior probability
@staticmethod
def mean_encode_blended(X_train, y_train, X_test, variable, target, weight_func):
"""
S_i represents an estimate of the probability of Y=1 given X=X_i
"""
X_train = X_train[[variable]].copy()
X_test = X_test[[variable]].copy()
if target is not None:
nf_name = '{}_pred_{}'.format(variable, target)
X_train['pred_temp'] = (y_train == target).astype(int)
else:
nf_name = '{}_pred'.format(variable)
X_train['pred_temp'] = y_train
# prior = n_Y / n_TR
prior = X_train['pred_temp'].mean()
# S_i['mean'] = n_iY/n_i and S_i['beta'] = lambda(n_i)
S_i = X_train.groupby(by=variable, axis=0)['pred_temp'].agg(mean="mean", beta="size")
S_i['beta'] = weight_func(S_i['beta'])
# Empirical Bayes Estimation: S_i = lambda(n_i)*n_iY/n_i + (1-lambda(n_i))*n_Y/n_TR
S_i[nf_name] = S_i['beta'] * S_i['mean'] + (1 - S_i['beta']) * prior
S_i.drop(['beta', 'mean'], axis=1, inplace=True)
nf_train = X_train.join(S_i, on=variable)[nf_name].values
nf_test = X_test.join(S_i, on=variable).fillna(prior, inplace=False)[nf_name].values
return nf_train, nf_test, prior, S_i
def get_params(self, deep=True):
return {
"cat_features": self.cat_features,
"target_type": self.target_type,
"cv": self.cv,
"k": self.k,
"f": self.f}
def set_params(self, **parameters):
for parameter, value in parameters.items():
setattr(self, parameter, value)
return selfPerform MeanEncoder() on ‘Blind_Make’, ‘Blind_Model’, ‘Blind_Submodel’
# High-cardinality categorical data
mean_encoder_feature = ['Blind_Make', 'Blind_Model', 'Blind_Submodel']
me = MeanEncoder(cat_features=mean_encoder_feature, cv=10, target_type='regression')
data_pre_feature4 = me.fit_transform(data_pre[mean_encoder_feature], data_pre["Claim_Amount"])
data_pre_feature4
Imbalanced Data
The data is highly imbalanced: more records contain zero claims than not. When designing your predictive model, you need to account for this.
There are a couple of ways to deal with imbalanced data.
- Resampling
- Over-sampling: SMOTE
- Under-sampling: Clustering, Tomek links
I built up-sampling and down-sampling functions to see whether they can improve the model.
from sklearn.utils import resample
from imblearn.pipeline import Pipeline
from imblearn.over_sampling import SMOTE
from imblearn.under_sampling import RandomUnderSampler
zero_label_num = len(data_pre_label[data_pre_label==0])
non_zero_label_num = len(data_pre_label[data_pre_label>0])
def upsampling(features, labels):
df = pd.concat([features, labels], axis=1)
# Separate majority and minority classes
df_majority = df[df.Claim_Amount==0]
df_minority = df[df.Claim_Amount!=0]
# Upsample minority class
df_minority_upsampled = resample(df_minority, replace=True, n_samples=df_majority.shape[0], random_state=914)
# Combine majority class with upsampled minority class
df_upsampled = pd.concat([df_majority, df_minority_upsampled])
# Display new class counts
print(
f"Non zero: {len(df_upsampled[df_upsampled.Claim_Amount>0])} \
Zero: {len(df_upsampled[df_upsampled.Claim_Amount==0])}")
return df_upsampled.drop('Claim_Amount', axis=1), df_upsampled.Claim_Amount
def downsampling(features, labels):
df = pd.concat([features, labels], axis=1)
# Separate majority and minority classes
df_majority = df[df.Claim_Amount==0]
df_minority = df[df.Claim_Amount!=0]
# Downsample majority class
df_majority_downsampled = resample(df_majority, replace=False, n_samples=df_minority.shape[0], random_state=411)
# Combine minority class with downsampled majority class
df_downsampled = pd.concat([df_majority_downsampled, df_minority])
# Display new class counts
print(
f"Non zero: {len(df_downsampled[df_downsampled.Claim_Amount>0])} \
Zero: {len(df_downsampled[df_downsampled.Claim_Amount==0])}")
return df_downsampled.drop('Claim_Amount', axis=1), df_downsampled.Claim_Amount
def smote_sampling(X, y):
smote = SMOTE(sampling_strategy="minority")
X, y = smote.fit_resample(X, y)
return X, yPut it all together!
def preprocessing_baseline(dataframe, use_upsampling=None, use_downsampling=None):
# Split into training and validation
data = dataframe.copy()
X_train, X_valid, y_train, y_valid = train_test_split(
data.drop("Claim_Amount", axis=1),
data.Claim_Amount,
test_size=0.15,
random_state=42,
stratify=(data.Claim_Amount!=0).astype(int))
if use_upsampling == True: X_train, y_train = upsampling(X_train, y_train)
if use_downsampling == True: X_train, y_train = downsampling(X_train, y_train)
# Define different datatype
int_features, float_features, object_features = type_of_col(data, label_col='Claim_Amount', show=False)
# ========================================
# Training
# ========================================
X_train.fillna(value=np.nan, inplace=True)
# Replace ? with np.nan
for col in object_features:
X_train.loc[:, col] = X_train.loc[:, col].replace(to_replace='?',value=np.nan)
X_train.loc[:, col] = X_train.loc[:, col].replace(to_replace='""',value=np.nan)
# Convert int to float
for col in int_features:
X_train[col] = X_train[col].astype('float64')
X_train = X_train.fillna(method='ffill')
full_transform = ColumnTransformer([
("num", StandardScaler(), int_features+float_features),
("cat", OneHotEncoder(), object_features)
])
X_train = full_transform.fit_transform(X_train)
# ========================================
# Validation
# ========================================
X_valid.fillna(value=np.nan, inplace=True)
# Replace ? with np.nan
for col in object_features:
X_valid.loc[:, col] = X_valid.loc[:, col].replace(to_replace='?',value=np.nan)
# Convert int to float
for col in int_features:
X_valid[col] = X_valid[col].astype('float64')
X_valid = X_valid.fillna(method='ffill')
X_valid = full_transform.transform(X_valid)
print(f"Size of training feature: {X_train.shape}\nSize of validation feature: {X_valid.shape}")
print(f"Size of training label: {y_train.shape}\nSize of validation label: {y_valid.shape}")
return X_train, X_valid, y_train, y_valid, full_transform
def preprocessing_v2(dataframe, use_upsampling=None, use_downsampling=None, use_blended=None):
# Split into training and validation
data = dataframe.copy()
X_train, X_valid, y_train, y_valid = train_test_split(
data.drop("Claim_Amount", axis=1),
data.Claim_Amount,
test_size=0.15,
random_state=42,
stratify=(data.Claim_Amount!=0).astype(int))
if use_upsampling == True: X_train, y_train = upsampling(X_train, y_train)
if use_downsampling == True: X_train, y_train = downsampling(X_train, y_train)
# Define different datatype
mean_encoder_feature = [
'Blind_Make', 'Blind_Model', 'Blind_Submodel']
num_feature = [
'Vehicle', 'Calendar_Year', 'Model_Year', 'Var1', 'Var2', 'Var3',
'Var4', 'Var5', 'Var6', 'Var7', 'Var8', 'NVVar1', 'NVVar2', 'NVVar3', 'NVVar4']
onehot_feature = [
'Cat1', 'Cat3', 'Cat6', 'Cat8', 'Cat9', 'Cat10', 'Cat11', 'Cat12', 'OrdCat', 'NVCat']
# ========================================
# Training
# ========================================
X_train = X_train.drop(['Row_ID', 'Household_ID', "Cat2", "Cat4", "Cat5", "Cat7"], axis=1)
X_train.fillna(value=np.nan, inplace=True)
# Replace ? and "" with np.nan
for col in onehot_feature:
X_train.loc[:, col] = X_train.loc[:, col].replace(to_replace='?',value=np.nan)
X_train.loc[:, col] = X_train.loc[:, col].replace(to_replace='""',value=np.nan)
# Convert int to float
for col in num_feature:
X_train[col] = X_train[col].astype('float64')
X_train = X_train.fillna(method='ffill')
full_transform = ColumnTransformer([
("num", MinMaxScaler(), num_feature),
("cat", OneHotEncoder(), onehot_feature),
("mean", MeanEncoder(cat_features=mean_encoder_feature, target_type="regression"), mean_encoder_feature)
])
X_train_final = full_transform.fit_transform(X_train, y_train)
# ========================================
# Validation
# ========================================
X_valid = X_valid.drop(['Row_ID', 'Household_ID', "Cat2", "Cat4", "Cat5", "Cat7"], axis=1)
X_valid.fillna(value=np.nan, inplace=True)
# Replace ? and "" with np.nan
for col in onehot_feature:
X_valid.loc[:, col] = X_valid.loc[:, col].replace(to_replace='?',value=np.nan)
X_valid.loc[:, col] = X_valid.loc[:, col].replace(to_replace='""',value=np.nan)
# Convert int to float
for col in num_feature:
X_valid[col] = X_valid[col].astype('float64')
X_valid = X_valid.fillna(method='ffill')
X_valid_final = full_transform.transform(X_valid)
print(f"Size of training data: {X_train_final.shape}\nSize of validation data: {X_valid_final.shape}")
print(f"Size of training label: {y_train.shape}\nSize of validation label: {y_valid.shape}")
return X_train_final, X_valid_final, y_train, y_valid, full_transform
def preprocessing_v3(dataframe, use_upsampling=None, use_downsampling=None, use_blended=None):
# Define different datatype
label = ["Claim_Amount"]
mean_encoder_feature = ['Blind_Make', 'Blind_Model']
num_feature = ['Var1', 'Var6', 'NVVar1', 'NVVar2', 'NVVar3', 'NVVar4']
onehot_feature = ['Cat1', 'Cat3', 'Cat6', 'Cat9', 'OrdCat', 'NVCat']
# Split into training and validation
data = dataframe.copy()
data = data[mean_encoder_feature+num_feature+onehot_feature+label]
X_train, X_valid, y_train, y_valid = train_test_split(
data.drop("Claim_Amount", axis=1),
data.Claim_Amount,
test_size=0.15,
random_state=42,
stratify=(data.Claim_Amount!=0).astype(int))
if use_upsampling == True: X_train, y_train = upsampling(X_train, y_train)
if use_downsampling == True: X_train, y_train = downsampling(X_train, y_train)
# ========================================
# Training
# ========================================
X_train.fillna(value=np.nan, inplace=True)
# Replace ? and "" with np.nan
for col in onehot_feature:
X_train.loc[:, col] = X_train.loc[:, col].replace(to_replace='?',value=np.nan)
X_train.loc[:, col] = X_train.loc[:, col].replace(to_replace='""',value=np.nan)
# Convert int to float
for col in num_feature:
X_train[col] = X_train[col].astype('float64')
X_train = X_train.fillna(method='ffill')
full_transform = ColumnTransformer([
("num", MinMaxScaler(), num_feature),
("cat", OneHotEncoder(), onehot_feature),
("mean", MeanEncoder(cat_features=mean_encoder_feature, target_type="regression"), mean_encoder_feature)
])
X_train_final = full_transform.fit_transform(X_train, y_train)
# ========================================
# Validation
# ========================================
X_valid.fillna(value=np.nan, inplace=True)
# Replace ? and "" with np.nan
for col in onehot_feature:
X_valid.loc[:, col] = X_valid.loc[:, col].replace(to_replace='?',value=np.nan)
X_valid.loc[:, col] = X_valid.loc[:, col].replace(to_replace='""',value=np.nan)
# Convert int to float
for col in num_feature:
X_valid[col] = X_valid[col].astype('float64')
X_valid = X_valid.fillna(method='ffill')
X_valid_final = full_transform.transform(X_valid)
print(f"Size of training data: {X_train_final.shape}\nSize of validation data: {X_valid_final.shape}")
print(f"Size of training label: {y_train.shape}\nSize of validation label: {y_valid.shape}")
return X_train_final, X_valid_final, y_train, y_valid, full_transformModelling
Tandem Model
Tandem is a two-stage regression method that can be used when various input data types are correlated, for example gene expression and methylation in drug response prediction. In the first stage it uses the upstream features (such as methylation) to predict the response variable (such as drug response), and in the second stage it uses the downstream features (such as gene expression) to predict the residuals of the first stage.
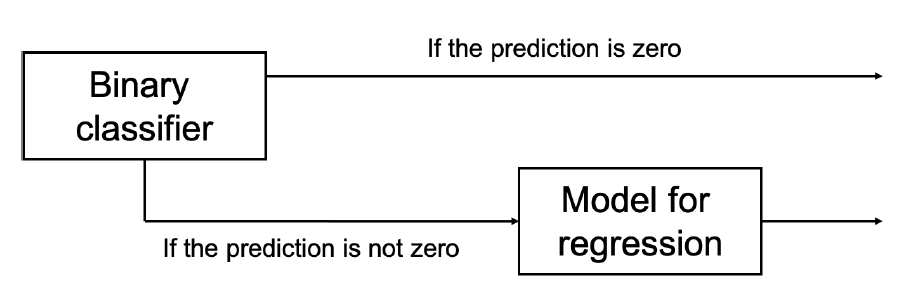
Pipeline
Performance using Single Model
You can see the problem as a regression problem where the variable to predict is continuous (the claimed amount in USD). The performance of the regression model will depend on the quality of the training data. I’ll compare the performance of the following models:
- Linear regression
- Ridge regression
- Random forests for regression
- Gradient tree boosting for regression
For each model, I’ll use grid search with at least three options for each parameter and report the performance measure over a validation set.
from sklearn.metrics import mean_squared_error
from sklearn.decomposition import TruncatedSVD
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Ridge
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import learning_curveLet’s split our data into training and validation dataset using preprocessing_v2() we built above.
X_train, X_valid, y_train, y_valid, _ = preprocessing_v2(data)Main Code
Linear Regression
RMSE: 274.5014
"""
Best params:
Origin: {'copy_X': True, 'fit_intercept': False, 'normalize': True}
Upsampling: {'copy_X': True, 'fit_intercept': True, 'normalize': False}
Downsampling: {'copy_X': True, 'fit_intercept': False, 'normalize': True}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'normalize': [True, False],
'fit_intercept': [True, False],
'copy_X': [True, False]
}
grid = GridSearchCV(LinearRegression(), cv=3, param_grid=param_grid, n_jobs=8)
grid.fit(X_train, y_train)
lr = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = lr.predict(X_valid)
rmse = np.sqrt(mean_squared_error(y_valid, y_pred))
print(f"RMSE: {round(rmse, 4)}")
else:
best_params = {
'copy_X': True, 'fit_intercept': False, 'normalize': True
}
lr = LinearRegression(**best_params)
lr.fit(X_train, y_train)
y_pred = lr.predict(X_valid)
rmse = np.sqrt(mean_squared_error(y_valid, y_pred))
print(f"RMSE: {round(rmse, 4)}")Ridge Regression
RMSE: 274.3249
"""
Best params:
Origin: {'alpha': 1.0, 'fit_intercept': True, 'normalize': True}
Upsampling: {'alpha': 1.0, 'fit_intercept': True, 'normalize': False}
Downsampling: {'alpha': 1.0, 'fit_intercept': True, 'normalize': False}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'alpha': [float(x) for x in np.linspace(0.1, 1.0, 10)],
'fit_intercept': [True, False],
'normalize':[True, False]
}
grid = GridSearchCV(Ridge(), cv=3, param_grid=param_grid, n_jobs=8)
grid.fit(X_train, y_train)
rr = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = rr.predict(X_valid)
rmse = np.sqrt(mean_squared_error(y_valid, y_pred))
print(f"RMSE: {round(rmse, 4)}")
else:
best_params = {
'alpha': 1.0, 'fit_intercept': True, 'normalize': True
}
rr = Ridge(**best_params)
rr.fit(X_train, y_train)
y_pred = rr.predict(X_valid)
rmse = np.sqrt(mean_squared_error(y_valid, y_pred))
print(f"RMSE: {round(rmse, 4)}")def pretty_print_coefficients(coefficients, names=None, sort=False):
if names == None:
names = ["X{}".format(x) for x in range(len(coefficients))]
lst = zip(coefficients, names)
if sort:
lst = sorted(lst, key=lambda x: -np.abs(x[0]))
return " + ".join("%s * %s" % (round(coef, 3), name) for coef, name in lst)
pretty_print_coefficients(rr.coef_, names=None, sort=False)'14.713 * X0 + -7.194 * X1 + 0.668 * X2 + 6.268 * X3 + 3.181 * X4 + 0.285 * X5 + 6.009 * X6 + 4.608 * X7 + -3.255 * X8 + 1.433 * X9 + -18.1 * X10 + -5.12 * X11 + 5.512 * X12 + -3.907 * X13 + -2.035 * X14 + -1.468 * X15 + 1.483 * X16 + -5.298 * X17 + 0.849 * X18 + -6.152 * X19 + -0.607 * X20 + 1.392 * X21 + 13.419 * X22 + -1.513 * X23 + -3.255 * X24 + 4.119 * X25 + -2.235 * X26 + -6.145 * X27 + -2.228 * X28 + -0.052 * X29 + -2.067 * X30 + -0.584 * X31 + 1.877 * X32 + 0.801 * X33 + -5.243 * X34 + -0.085 * X35 + 1.982 * X36 + -0.864 * X37 + -4.318 * X38 + -0.041 * X39 + 0.041 * X40 + 0.981 * X41 + -0.338 * X42 + -3.315 * X43 + 0.591 * X44 + 0.117 * X45 + 0.963 * X46 + 0.254 * X47 + -3.111 * X48 + -1.329 * X49 + 42.711 * X50 + -0.511 * X51 + 1.54 * X52 + 0.599 * X53 + -3.295 * X54 + -1.784 * X55 + 58.729 * X56 + -0.35 * X57 + -7.959 * X58 + 0.052 * X59 + 0.335 * X60 + -1.077 * X61 + -21.379 * X62 + 6.138 * X63 + -0.224 * X64 + 11.816 * X65 + -1.387 * X66 + 3.474 * X67 + -3.939 * X68 + -10.15 * X69 + -7.045 * X70 + -15.198 * X71 + -1.191 * X72 + -0.275 * X73 + -2.104 * X74 + 0.844 * X75 + 1.142 * X76 + -0.481 * X77 + 0.041 * X78 + -0.005 * X79 + -0.005 * X80'Random Forest
RMSE: 274.4148
"""
Best params:
Origin: {'max_depth': 3, 'min_samples_split': 10, 'n_estimators': 100}
Upsampling: {'max_depth': 5, 'min_samples_split': 5, 'n_estimators': 100}
Downsampling: {'max_depth': 5, 'min_samples_split': 5, 'n_estimators': 200}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'n_estimators': [int(x) for x in np.linspace(100, 300, 3)],
'max_depth': [int(x) for x in range(3, 6)],
'min_samples_split': [2, 5, 10],
# 'min_samples_leaf': [1, 2, 4],
# 'bootstrap': [True, False]
}
grid = GridSearchCV(RandomForestRegressor(), cv=3, param_grid=param_grid, n_jobs=8)
grid.fit(X_train, y_train)
rfr = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = rfr.predict(X_valid)
rmse = np.sqrt(mean_squared_error(y_valid, y_pred))
print(f"RMSE: {round(rmse, 4)}")
else:
best_params = {
'max_depth': 3, 'min_samples_split': 10, 'n_estimators': 100
}
rfr = RandomForestRegressor(**best_params)
rfr.fit(X_train, y_train)
y_pred = rfr.predict(X_valid)
rmse = np.sqrt(mean_squared_error(y_valid, y_pred))
print(f"RMSE: {round(rmse, 4)}")Gradient Tree Boosting
RMSE: 274.5765
"""
Best params:
Origin: {'max_depth': 3, 'min_samples_split': 0.5, 'n_estimators': 200}
Upsampling: {'max_depth': 5, 'min_samples_split': 0.1, 'n_estimators': 500}
Downsampling: {'max_depth': 3, 'min_samples_split': 0.5, 'n_estimators': 200}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'n_estimators': [int(x) for x in np.linspace(200, 400, 3)],
'max_depth': [int(x) for x in range(3, 6)],
"min_samples_split": np.linspace(0.1, 0.5, 12),
# 'learning_rate': [0.1, 0.01, 0.001],
# 'min_samples_leaf': np.linspace(0.1, 0.5, 12),
# "subsample": [0.5, 0.618, 0.8, 0.85, 0.9, 0.95, 1.0],
# 'max_features': ["log2","sqrt"]
}
grid = GridSearchCV(GradientBoostingRegressor(), cv=3, param_grid=param_grid, n_jobs=8)
grid.fit(X_train, y_train)
gbr = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = gbr.predict(X_valid)
rmse = np.sqrt(mean_squared_error(y_valid, y_pred))
print(f"RMSE: {round(rmse, 4)}")
else:
best_params = {
'max_depth': 3, 'min_samples_split': 0.5, 'n_estimators': 200
}
gbr = GradientBoostingRegressor(**best_params)
gbr.fit(X_train, y_train)
y_pred = gbr.predict(X_valid)
rmse = np.sqrt(mean_squared_error(y_valid, y_pred))
print(f"RMSE: {round(rmse, 4)}")Performance
from IPython.display import HTML, display
report = [
["", "RMSE (original)", "RMSE (up-sampling)", "RMSE (down-sampling)"],
["Linear Regression", 204.9581, 214.4630, 214.3920],
["Ridge Regression", 204.6595, 214.4686, 214.3732],
["Random Forest", 204.6774, 221.9525, 233.0449],
["Gradient Tree Boosting", 205.4188, 230.6640, 219.3338]
]
display(HTML(
'<table><tr>{}</tr></table>'.format(
'</tr><tr>'.join(
'<td>{}</td>'.format('</td><td>'.join(str(_) for _ in row)) for row in report))))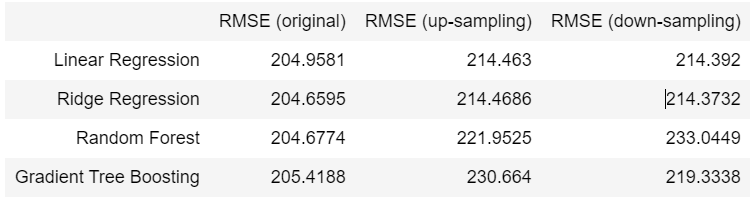
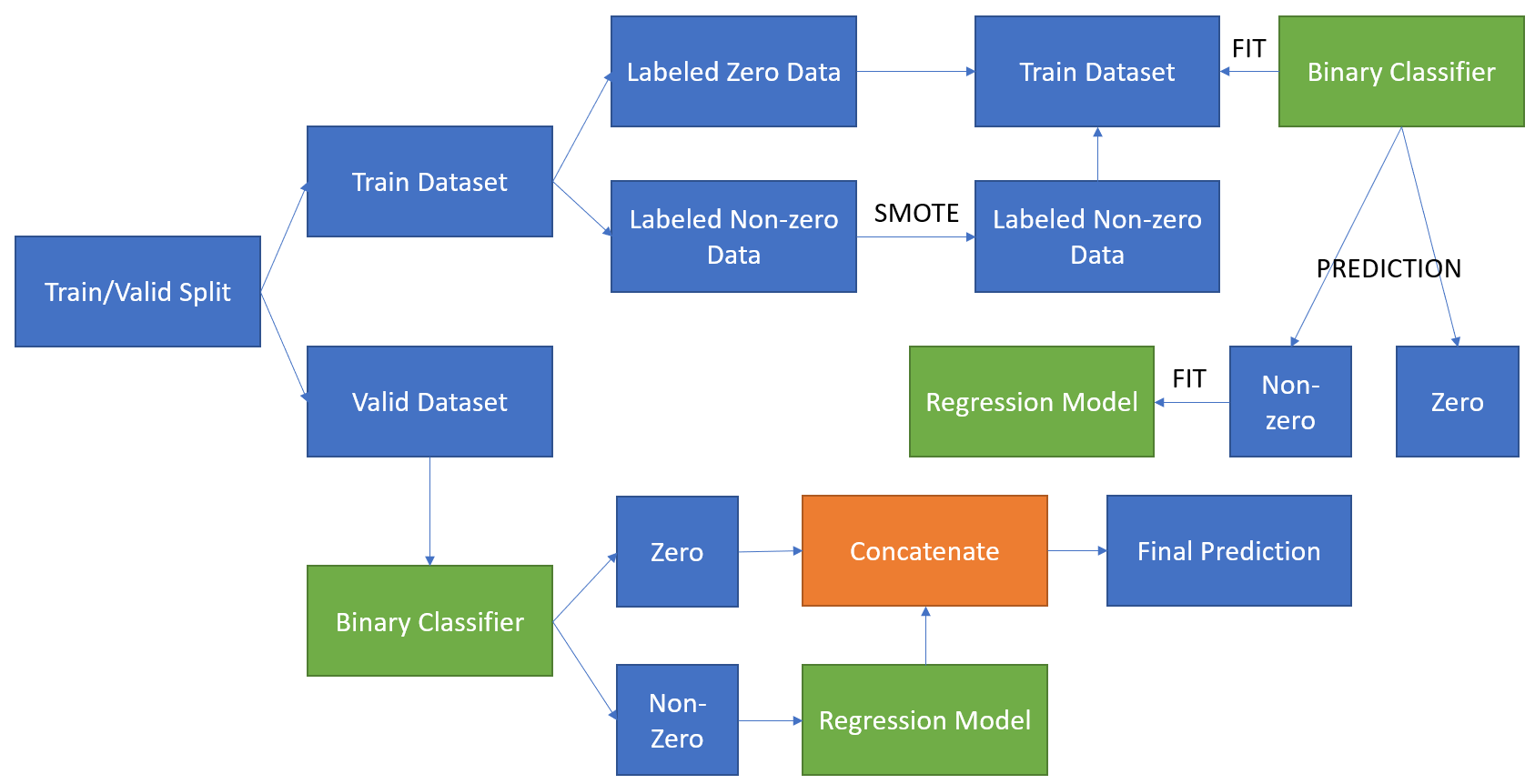
Performance using a combination of two models
In this section, I will build a prediction model based on two separate models in tandem (one after the other). The first model will be a binary classifier that will tell whether the claim was zero or different from zero. I will compare the following classifiers: random forests for classification and gradient boosting for classification.
As usual, load in required libraries.
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_reportAnd split it into training and validation dataset using preprocessing_v2().
X_train, X_valid, y_train, y_valid, full_transform = preprocessing_v2(data)
y_train_binary = (y_train != 0.0).astype(int)
y_valid_binary = (y_valid != 0.0).astype(int)Second Model
Random Forest
"""
Best params:
Origin: {'max_depth': 3, 'min_samples_split': 2, 'n_estimators': 100}
Upsampling: {'max_depth': 9, 'min_samples_split': 7, 'n_estimators': 300}
Downsampling: {'max_depth': 9, 'min_samples_split': 7, 'n_estimators': 300}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'n_estimators': [int(x) for x in np.linspace(300, 500, 3)],
'max_depth': [int(x) for x in range(5, 10)],
'min_samples_split': [7, 9, 10]
}
grid = GridSearchCV(RandomForestClassifier(), cv=3, param_grid=param_grid, n_jobs=8, scoring="f1")
grid.fit(X_train, y_train_binary)
rfc = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = rfc.predict(X_valid)
else:
best_params = {
'max_depth': 3, 'min_samples_split': 2, 'n_estimators': 100
}
rfc = RandomForestClassifier(**best_params)
rfc.fit(X_train, y_train_binary)
y_pred = rfc.predict(X_valid)
print(classification_report(y_valid_binary, y_pred))
print(confusion_matrix(y_valid_binary, y_pred))Gradient Boosting
"""
Best params:
Origin: {'max_depth': 3, 'min_samples_split': 0.30000000000000004, 'n_estimators': 300}
Upsampling: {'max_depth': 5, 'min_samples_split': 0.1, 'n_estimators': 400}
Downsampling: {'max_depth': 5, 'min_samples_split': 0.1, 'n_estimators': 300}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'n_estimators': [int(x) for x in np.linspace(200, 400, 3)],
'max_depth': [int(x) for x in range(3, 6)],
"min_samples_split": np.linspace(0.1, 0.5, 3),
# 'learning_rate': [0.1, 0.01, 0.001],
# 'min_samples_leaf': np.linspace(0.1, 0.5, 12),
# "subsample": [0.5, 0.618, 0.8, 0.85, 0.9, 0.95, 1.0],
# 'max_features': ["log2","sqrt"]
}
grid = GridSearchCV(GradientBoostingClassifier(), cv=3, param_grid=param_grid, n_jobs=8, scoring="f1")
grid.fit(X_train, y_train_binary)
gbc = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = gbc.predict(X_valid)
else:
best_params = {
'max_depth': 3, 'min_samples_split': 0.30000000000000004, 'n_estimators': 300
}
gbc = GradientBoostingClassifier(**best_params)
gbc.fit(X_train, y_train_binary)
y_pred = gbc.predict(X_valid)
print(classification_report(y_valid_binary, y_pred))
print(confusion_matrix(y_valid_binary, y_pred))Put these two models as second model, and combine with primary model. For the second model, if the claim was different from zero, train a regression model to predict the actual value of the claim.
This time, I’ll put all code together inside a code block.
"""
Best params:
Origin: {'copy_X': True, 'fit_intercept': False, 'normalize': True}
Upsampling: {'copy_X': True, 'fit_intercept': True, 'normalize': False}
Downsampling: {'copy_X': True, 'fit_intercept': True, 'normalize': True}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'normalize': [True, False],
'fit_intercept': [True, False],
'copy_X': [True, False]
}
grid = GridSearchCV(LinearRegression(), cv=3, param_grid=param_grid, n_jobs=8)
grid.fit(X_train[np.where(y_train != 0)], y_train[y_train != 0])
lr_2 = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = lr_2.predict(X_valid[np.where(y_valid != 0)])
rmse = np.sqrt(mean_squared_error(y_valid[y_valid != 0], y_pred))
print(f"RMSE: {round(rmse, 4)}")
else:
best_params = {
'copy_X': True, 'fit_intercept': True, 'normalize': True
}
lr_2 = LinearRegression(**best_params)
lr_2.fit(X_train[np.where(y_train != 0)], y_train[y_train != 0])
y_pred = lr_2.predict(X_valid[np.where(y_valid != 0)])
rmse = np.sqrt(mean_squared_error(y_valid[y_valid != 0], y_pred))
print(f"RMSE: {round(rmse, 4)}")
"""
Best params:
Origin: {'alpha': 1.0, 'fit_intercept': True, 'normalize': True}
Upsampling: {'alpha': 0.1, 'fit_intercept': True, 'normalize': True}
Downsampling: {'alpha': 1.0, 'fit_intercept': True, 'normalize': True}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'alpha': [float(x) for x in np.linspace(0.1, 1.0, 10)],
'fit_intercept': [True, False],
'normalize':[True, False]
}
grid = GridSearchCV(Ridge(), cv=3, param_grid=param_grid, n_jobs=8)
grid.fit(X_train[np.where(y_train != 0)], y_train[y_train != 0])
rr_2 = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = rr_2.predict(X_valid[np.where(y_valid != 0)])
rmse = np.sqrt(mean_squared_error(y_valid[y_valid != 0], y_pred))
print(f"RMSE: {round(rmse, 4)}")
else:
best_params = {
'alpha': 1.0, 'fit_intercept': True, 'normalize': True
}
rr_2 = Ridge(**best_params)
rr_2.fit(X_train[np.where(y_train != 0)], y_train[y_train != 0])
y_pred = rr_2.predict(X_valid[np.where(y_valid != 0)])
rmse = np.sqrt(mean_squared_error(y_valid[y_valid != 0], y_pred))
print(f"RMSE: {round(rmse, 4)}")
"""
Best params:
Origin: {'max_depth': 3, 'min_samples_split': 2, 'n_estimators': 100}
Upsampling: {'max_depth': 5, 'min_samples_split': 2, 'n_estimators': 300}
Downsampling: {'max_depth': 3, 'min_samples_split': 2, 'n_estimators': 300}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'n_estimators': [int(x) for x in np.linspace(100, 300, 3)],
'max_depth': [int(x) for x in range(3, 6)],
'min_samples_split': [2, 5, 10],
# 'min_samples_leaf': [1, 2, 4],
# 'bootstrap': [True, False]
}
grid = GridSearchCV(RandomForestRegressor(), cv=3, param_grid=param_grid, n_jobs=8)
grid.fit(X_train[np.where(y_train != 0)], y_train[y_train != 0])
rfr_2 = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = rfr_2.predict(X_valid[np.where(y_valid != 0)])
rmse = np.sqrt(mean_squared_error(y_valid[y_valid != 0], y_pred))
print(f"RMSE: {round(rmse, 4)}")
else:
best_params = {
'max_depth': 3, 'min_samples_split': 2, 'n_estimators': 100
}
rfr_2 = RandomForestRegressor(**best_params)
rfr_2.fit(X_train[np.where(y_train != 0)], y_train[y_train != 0])
y_pred = rfr_2.predict(X_valid[np.where(y_valid != 0)])
rmse = np.sqrt(mean_squared_error(y_valid[y_valid != 0], y_pred))
print(f"RMSE: {round(rmse, 4)}")
"""
Best params:
Origin: {'max_depth': 3, 'min_samples_split': 0.5, 'n_estimators': 200}
Upsampling: {'max_depth': 5, 'min_samples_split': 0.1, 'n_estimators': 400}
Downsampling: {'max_depth': 3, 'min_samples_split': 0.5, 'n_estimators': 200}
"""
USE_GRID = False
if USE_GRID:
param_grid = {
'n_estimators': [int(x) for x in np.linspace(200, 400, 3)],
'max_depth': [int(x) for x in range(3, 6)],
"min_samples_split": np.linspace(0.1, 0.5, 12),
# 'learning_rate': [0.1, 0.01, 0.001],
# 'min_samples_leaf': np.linspace(0.1, 0.5, 12),
# "subsample": [0.5, 0.618, 0.8, 0.85, 0.9, 0.95, 1.0],
# 'max_features': ["log2","sqrt"]
}
grid = GridSearchCV(GradientBoostingRegressor(), cv=3, param_grid=param_grid, n_jobs=8)
grid.fit(X_train[np.where(y_train != 0)], y_train[y_train != 0])
gbr_2 = grid.best_estimator_
print(f"Best params: {grid.best_params_}")
y_pred = gbr_2.predict(X_valid[np.where(y_valid != 0)])
rmse = np.sqrt(mean_squared_error(y_valid[y_valid != 0], y_pred))
print(f"RMSE: {round(rmse, 4)}")
else:
best_params = {
'max_depth': 3, 'min_samples_split': 0.5, 'n_estimators': 200
}
gbr_2 = GradientBoostingRegressor(**best_params)
gbr_2.fit(X_train[np.where(y_train != 0)], y_train[y_train != 0])
y_pred = gbr_2.predict(X_valid[np.where(y_valid != 0)])
rmse = np.sqrt(mean_squared_error(y_valid[y_valid != 0], y_pred))
print(f"RMSE: {round(rmse, 4)}")Use the tandem model built from before, for predicting in the same validation data used in the beginning, and report the performance.
RandomForestClassifier + LinearRegression
final_prediction = []
first_prediction = rfc.predict(X_valid)
for i, pred in enumerate(first_prediction):
if pred == 0:
final_prediction.append(0)
else:
non_zero_prediction = lr_2.predict(X_valid[i].reshape(1, -1))
final_prediction.append(non_zero_prediction[0])
round(np.sqrt(mean_squared_error(y_valid, final_prediction)), 4)RandomForestClassifier + Ridge
final_prediction = []
first_prediction = rfc.predict(X_valid)
for i, pred in enumerate(first_prediction):
if pred == 0:
final_prediction.append(0)
else:
non_zero_prediction = rr_2.predict(X_valid[i].reshape(1, -1))
final_prediction.append(non_zero_prediction[0])
round(np.sqrt(mean_squared_error(y_valid, final_prediction)), 4)RandomForestClassifier + RandomForestRegressor
final_prediction = []
first_prediction = rfc.predict(X_valid)
for i, pred in enumerate(first_prediction):
if pred == 0:
final_prediction.append(0)
else:
non_zero_prediction = rfr_2.predict(X_valid[i].reshape(1, -1))
final_prediction.append(non_zero_prediction[0])
round(np.sqrt(mean_squared_error(y_valid, final_prediction)), 4)RandomForestClassifier + GradientBoostinRegressor
final_prediction = []
first_prediction = rfc.predict(X_valid)
for i, pred in enumerate(first_prediction):
if pred == 0:
final_prediction.append(0)
else:
non_zero_prediction = gbr_2.predict(X_valid[i].reshape(1, -1))
final_prediction.append(non_zero_prediction[0])
round(np.sqrt(mean_squared_error(y_valid, final_prediction)), 4)GradientBoostingClassifier + LinearRegression
final_prediction = []
first_prediction = gbc.predict(X_valid)
for i, pred in enumerate(first_prediction):
if pred == 0:
final_prediction.append(0)
else:
non_zero_prediction = lr_2.predict(X_valid[i].reshape(1, -1))
final_prediction.append(non_zero_prediction[0])
round(np.sqrt(mean_squared_error(y_valid, final_prediction)), 4)GradientBoostingClassifier + Ridge
final_prediction = []
first_prediction = gbc.predict(X_valid)
for i, pred in enumerate(first_prediction):
if pred == 0:
final_prediction.append(0)
else:
non_zero_prediction = rr_2.predict(X_valid[i].reshape(1, -1))
final_prediction.append(non_zero_prediction[0])
round(np.sqrt(mean_squared_error(y_valid, final_prediction)), 4)GradientBoostingClassifier + RandomForestRegressor
final_prediction = []
first_prediction = gbc.predict(X_valid)
for i, pred in enumerate(first_prediction):
if pred == 0:
final_prediction.append(0)
else:
non_zero_prediction = rfr_2.predict(X_valid[i].reshape(1, -1))
final_prediction.append(non_zero_prediction[0])
round(np.sqrt(mean_squared_error(y_valid, final_prediction)), 4)GradientBoostingClassifier + GradientBoostingRegressor
final_prediction = []
first_prediction = gbc.predict(X_valid)
for i, pred in enumerate(first_prediction):
if pred == 0:
final_prediction.append(0)
else:
non_zero_prediction = gbr_2.predict(X_valid[i].reshape(1, -1))
final_prediction.append(non_zero_prediction[0])
round(np.sqrt(mean_squared_error(y_valid, final_prediction)), 4)Finally, performance of every models come out!
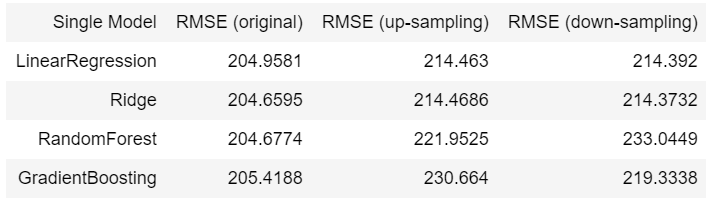
Single Model:
Tandem Model:
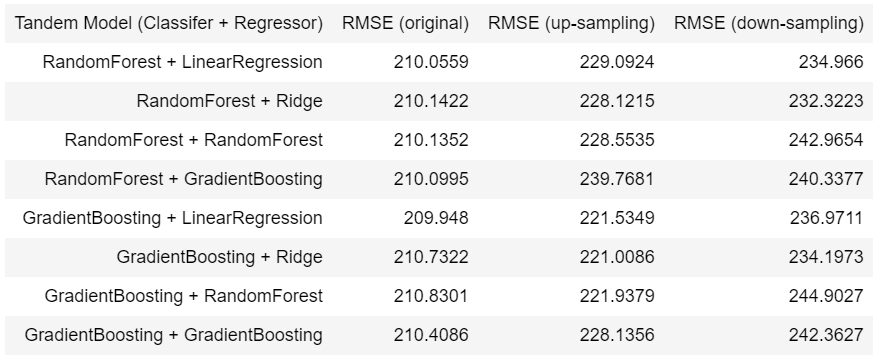
The best model from step 2 is using LinearRegression without any over sampling and down sampling technique. And the best model from step 3 is using RandomForest + LinearRegression without any over sampling and down sampling. Mean squared error for the best model from step 2 and 3 are 214.463 and 210.0559, respectively.
Conclusion
- For the single regression models, up-sampling and down-sampling technique do not give advantages at all. Actually, they even have higher mean squared error for those four models. Furthermore, we can see that when using resampling methods, mean square error of tree-based models are slightly higher than linear models.
- I utilised different data preprocessing approaches to encode categorical features, e.g.
preprocessing_baseline()andpreprocessing_v2(), such asOneHotEncoder()andMeanEncoder(). In baseline preprocessor, I usedOneHotEncoder()only to transform the categorical feature. But I found thatOneHotEncoder()would produce high sparsity matrix when there’s many categories. Therefore, I subclass aMeanEncoder()in sklearn to deal with high-cardinality categorical features. After the experiment, usingMeanEncoder()moderately improves the performance of the models. - For the tandem models, I trained the models in two different ways. First, I built a binary classifier, and collected the prediction which is not zero. Next, I fed them into regression model to get the final predicitons. Second, I built a binary classifier as the one before, and after that I selected only non-zero labels in original dataset to feed into regression model. When I done training these two methods, the second one performs better than the first one, so I decided to use the second training method as my pipeline.
- When comparing single models with tandem models, it can see that single models is better than tandem models whether or not sampling methods are used. Single models’ mean square error are less than tandem models’ by 2.85% in average. However, if we must utilise sampling method to solve imbalanced label problem, it can be seen that over sampling practically outperforms down sampling.

